Results for 6c26
EMD-7336
Membrane mimetics: micelle; Digitonin Note: More manual adjustment needed because of the noise level; there is missleading density in slices 330-350
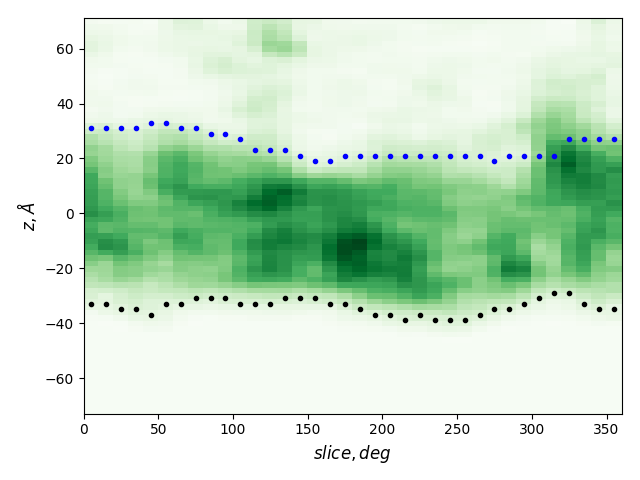
Cross-sections at different angles
green: density, gray dots: atoms from the all-atom model, lines: membrane boundaries defined by TMDET
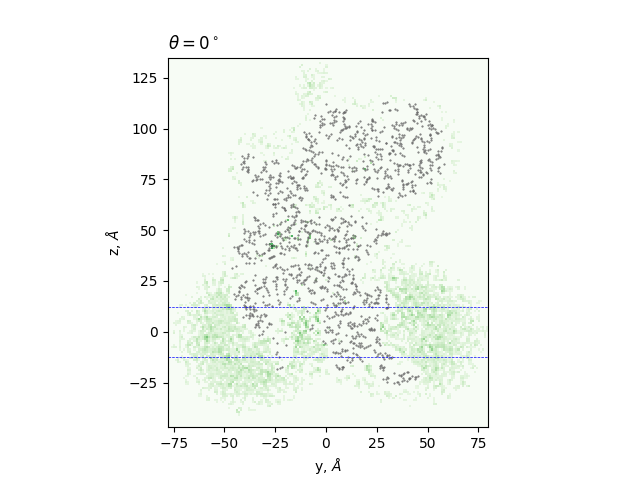
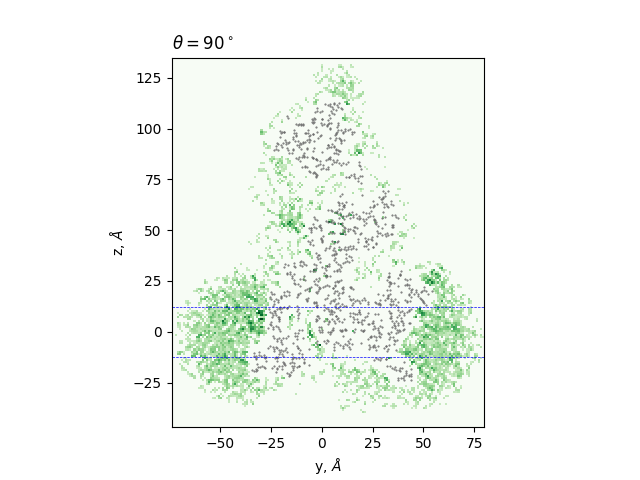
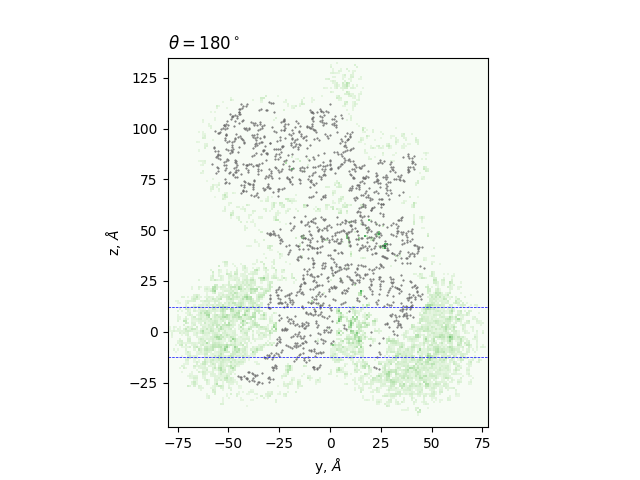
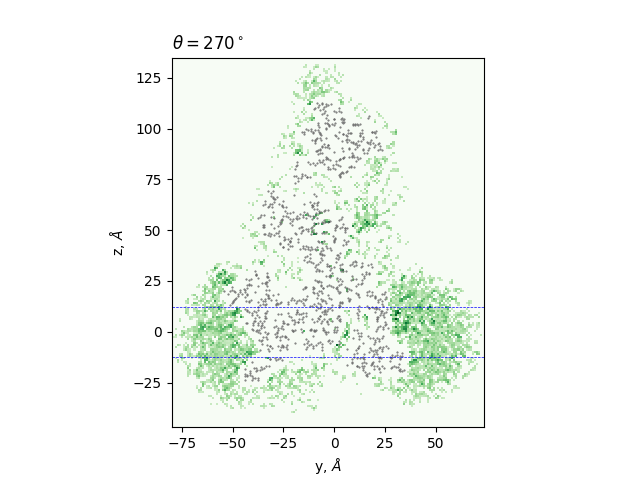
The determined edges at each slice and their comparison to TMDET
dots: edges of the gradient, triangles: outliers,
dashed and solid lines: membrane boundaries defined by TMDET and MemBlob ( mean(Zi) decreased by the thickness of the interface region (8 Å))
[ Download results
Resubmit ]
Manual settings applied
110-130 end1 average 100 140
190-310 end1 around 20 5
320-350 end1 around 28 2
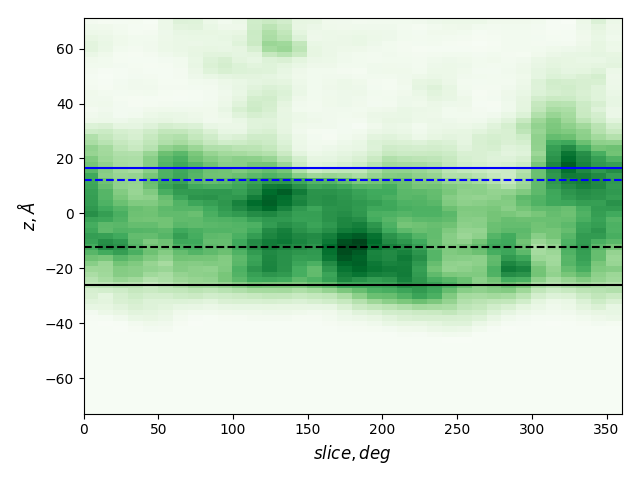
Boundary values (averages for MemBlob)
| Z (PDBTM) | Z (MemBlob) | Z - 8 | |
| End1 | 12.25 | 24.61 | 16.61 |
| End0 | -12.25 | -34.00 | -26.00 |
Mapping the results to the structure
You can move/scale/rotate the structure.
Blue: a.a. facing to bulk water;
magenta: interface region;
green: a.a. interacting with the hydrophobic membrane core;
yellow: burried a.a.
red: a.a. with an invisible side chain and non-protein molecules, for which DSSP cannot return SASA
[ Download results
Resubmit ]

