What is MemBlob?
MemBlob implements the first algorithm ever to determine the membrane boundaries of transmembrane proteins based on a larger set of experimental data, namely electron densities. The membrane environment used for structure determination cannot be resolved at the atomic level, but it produces electron density, which can be utilized to dissect the membrane embedded part of the protein.
INPUT:
- An electron density map in MRC format (e.g. from rutgers.edu)
- The corresponding all-atom structure of the protein (e.g. from RCSB)
- The membrane region prediction from PDBTM or TMDET
OUTPUT:
- A pdb file with B-factors labeling the positions of atoms:
- on the surface
- in the water phase
- in the membrane interface region
- in the hydrophobic core
- buried inside the protein
- Other files, such as imageof slices and projections
IMPORTANCE:
- The membrane region is defined based on experiments
- The membrane is not defined as a slab, but follows the shape of the membrane environment
- You can use this data for setting energy tags in algorithms for transmembrane protein (re)design, in studies of protein/drug interactions, etc.
PIPELINE:
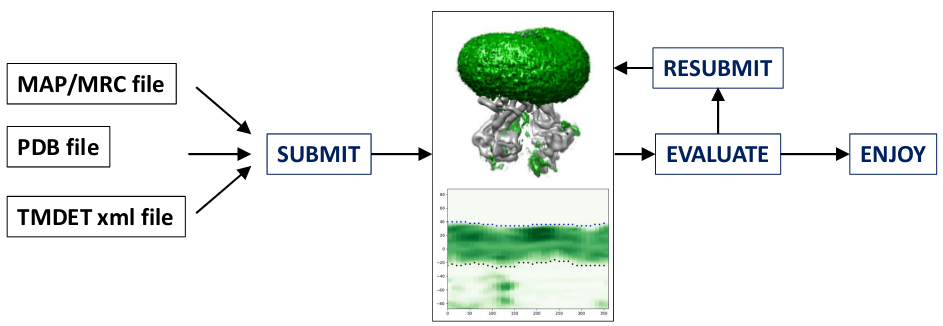
Notice: Online Calculation Tool Unavailable
Due to recent technological advancements, our online calculation tool (Submit Page) is no longer operational.
If you need to perform MemBlob calculations, please contact us to accelerate the process.
Thank you for your patience and understanding!
DATABASE:
We ran our pipline on structures determined by cryo-EM at a resolution better than 4 Å. Structures, corresponding electron density maps, and TMDET XML files were downloaded from RCSB, EMD, and TMDET respectively (July, 2018).
PUBLICATION:
MemBlob database and server for identifying transmembrane regions using cryo-EM maps
Bianka Farkas1,2,3,#, Georgina Csizmadia1,2,#, Eszter Katona1,4, Gábor E. Tusnády5, Tamás Hegedűs1,2,*
1Department of Biophysics and Radiation Biology, Semmelweis University, Budapest, HU,
2MTA-SE Molecular Biophysics Research Group, Hungarian Academy of Sciences, Budapest, HU,
3Faculty of Information Technology and Bionics, Pázmány Péter Catholic University, Budapest, HU,
4University College London, London, UK,
5"Momentum" Membrane Protein Bioinformatics Research Group, Institute of Enzymology, RCNS, Hungarian Academy of Sciences
2MTA-SE Molecular Biophysics Research Group, Hungarian Academy of Sciences, Budapest, HU,
3Faculty of Information Technology and Bionics, Pázmány Péter Catholic University, Budapest, HU,
4University College London, London, UK,
5"Momentum" Membrane Protein Bioinformatics Research Group, Institute of Enzymology, RCNS, Hungarian Academy of Sciences
Bioinformatics, btz539, https://doi.org/10.1093/bioinformatics/btz539
Thanks for the suggestions and critical notes to Hedvig Tordai.